I have already demonstrated how to create a knowledge graph out of a Wikipedia page. However, since the post got a lot of attention, I’ve decided to explore other domains where using NLP techniques to construct a knowledge graph makes sense. In my opinion, the biomedical field is a prime example where representing the data as a graph makes sense as you are often analyzing interactions and relations between genes, diseases, drugs, proteins, and more.
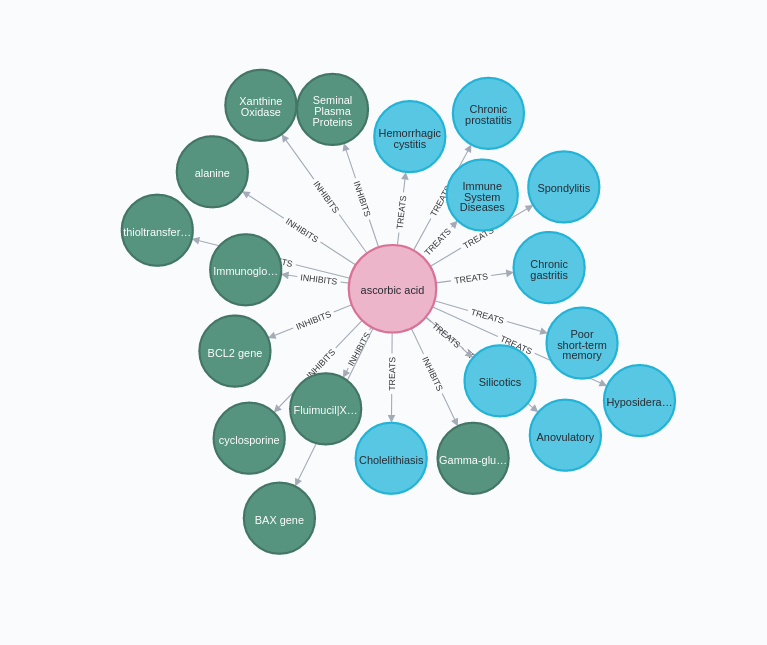
In the above visualization, we have ascorbic acid, also known as vitamin C, and some of its relations to other concepts. For example, it shows that vitamin C could be used to treat chronic gastritis.
Now, you could have a team of domain experts map all of those connections between drugs, diseases, and other biomedical concepts for you. But, unfortunately, not many of us can afford to hire a team of medical doctors to do the work for us. In that case, we can resort to using NLP techniques to extract those relationships automatically. The good part is that we can use an NLP pipeline to read all of the research papers out there, and the bad part is that not all obtained results will be perfect. However, given that I don’t have a team of scientists ready at my side to extract relations manually, I will resort to using NLP techniques to construct a biomedical knowledge graph of my own.
I will be using a single research paper in this blog post to walk you through all the steps required to construct a biomedical knowledge graph – Tissue Engineering of Skin Regeneration and Hair Growth.
The paper was written by Mohammadreza Ahmadi. The PDF version of the article is available under the CC0 1.0 license. We will go through the following steps to construct a knowledge graph:
- Reading a PDF document with OCR
- Text preprocessing
- Biomedical concept recognition and linking
- Relation extraction
- External database enrichment
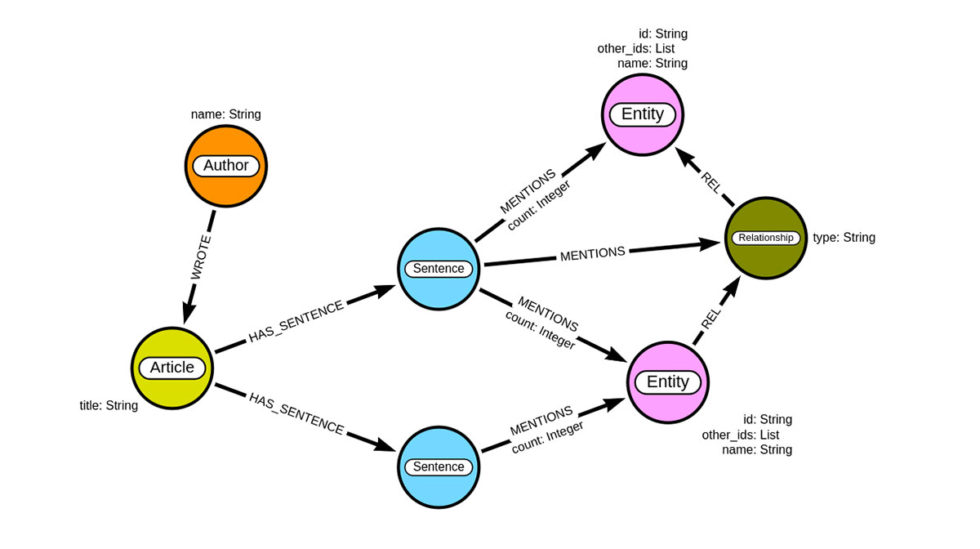
By the end of this post, you will construct a graph with the following schema.
We will be using Neo4j, a graph database that features the labeled property graph model, to store our graph. Each article can have one or more authors. We will split the article content into sentences and use NLP to extract both medical entities and their relationships. It might be a bit counter-intuitive that we will store the relations between entities as intermediate nodes instead of relationships. The critical factor behind this decision is that we want to have an audit trail of the source text from which the relation was extracted. With the labeled property graph model, you can’t have a relationship pointing to another relationship. For this reason, we refactor the connection between medical concepts into an intermediate node. This will also allow a domain expert to evaluate if a relation was correctly extracted or not.
Along the way, I will also demonstrate applications of using the constructed graph to search and analyze stored information.
Let’s dive right into it!
If this in-depth educational content is useful for you, subscribe to our AI research mailing list to be alerted when we release new material.
Reading a PDF document with OCR
As mentioned, the PDF version of the research paper is accessible to the public under the CC0 1.0 license, which means we can easily download it with Python. We will be using the pytesseract library to extract text from the PDF. As far as I know, the pytesseract library is one of the more popular libraries for OCR. If you want to follow along with code examples, I have prepared a Google Colab notebook, so you don’t have to copy-paste the code yourself.
importrequestsimportpdf2imageimportpytesseractdoc=pdf2image.convert_from_bytes(pdf.content)# Get the article textarticle=[]forpage_number, page_datainenumerate(doc):txt=pytesseract.image_to_string(page_data).encode("utf-8")# Sixth page are only referencesifpage_number <6:article.append(txt.decode("utf-8"))article_txt=" ".join(article)
Text preprocessing
Now that we have the article content available, we will go ahead and remove section titles and figure descriptions from the text. Next, we will split the text into sentences.
importnltknltk.download('punkt')defclean_text(text):"""Remove section titles and figure descriptions from text"""clean="\n".join([rowforrowintext.split("\n")if(len(row.split(" "))) >3andnot(row.startswith("(a)"))andnotrow.startswith("Figure")])returncleantext=article_txt.split("INTRODUCTION")[1]ctext=clean_text(text)sentences=nltk.tokenize.sent_tokenize(ctext)
Biomedical named entity linking
Now comes the exciting part. For those new to NLP and named entity recognition and linking, let’s start with some basics. Named entity recognition techniques are used to detect relevant entities or concepts in the text. For example, in the biomedical domain, we want to identify various genes, drugs, diseases, and other concepts in the text.
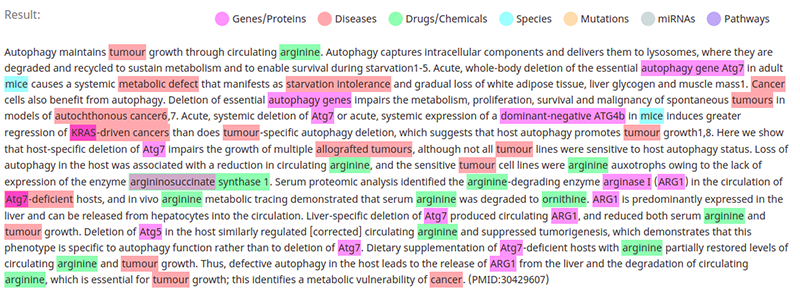
In this example, the NLP model identified genes, diseases, drugs, species, mutations, and pathways in the text. As mentioned, this process is called named entity recognition. An upgrade to the named entity recognition is the so-called named entity linking. The named entity linking technique detects relevant concepts in the text and tries to map them to the target knowledge base. In the biomedical domain, some of the target knowledge bases are:
Why would we want to link medical entities to a target knowledge base? The primary reason is that it helps us deal with entity disambiguation. For example, we don’t want separate entities in the graph representing ascorbic acid and vitamin C as domain experts can tell you those are the same thing. The secondary reason is that by mapping concepts to a target knowledge base, we can enrich our graph model by fetching information about the mapped concepts from the target knowledge base. If we use the ascorbic acid example again, we could easily fetch additional information from the CHEBI database if we already know its CHEBI id.
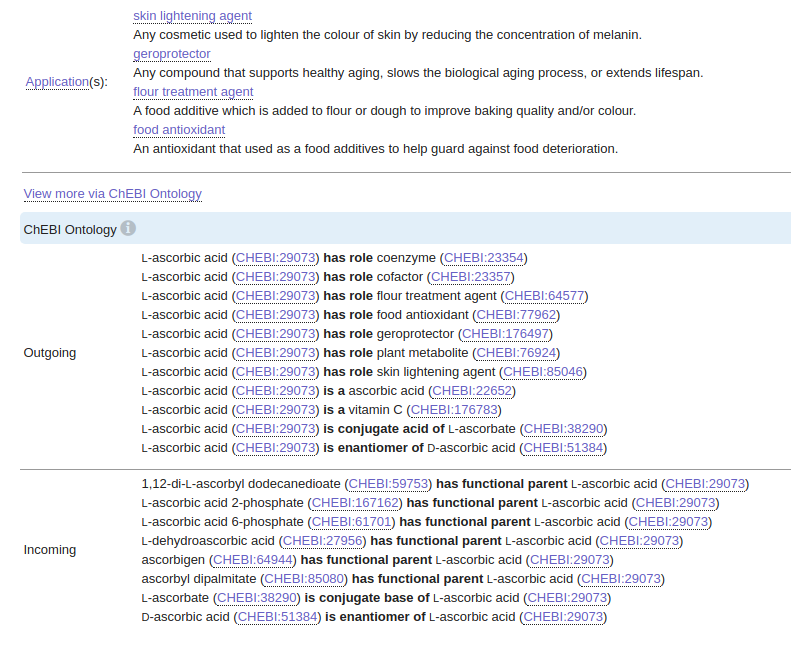
I’ve been looking for a decent open-source pre-trained biomedical named entity linking for some time. Lots of NLP models focus on extracting only a specific subset of medical concepts like genes or diseases. It is even rarer to find a model that detects most medical concepts and links them to a target knowledge base. Luckily I’ve stumbled upon BERN[1], a neural biomedical entity recognition and multi-type normalization tool. If I understand correctly, it is a fine-tuned BioBert model with various named entity linking models integrated for mapping concepts to biomedical target knowledge bases. Not only that, but they also provide a free REST endpoint, so we don’t have to deal with the headache of getting the dependencies and the model to work. The biomedical named entity recognition visualization I’ve used above was created using the BERN model, so we know it detects genes, diseases, drugs, species, mutations, and pathways in the text.
Unfortunately, the BERN model does not assign target knowledge base ids to all concepts. So I’ve prepared a script that first looks if a distinct id is given for a concept, and if it is not, it will use the entity name as the id. We will also compute the sha256 of the text of sentences to identify specific sentences easier later when we will be doing relation extraction.
importhashlib"""Biomedical entity linking API"""returnrequests.post(url, data={'sample_text': text}).json()entity_list=[]# The last sentence is invalidforsinsentences[:-1]:entity_list.append(query_raw(s))parsed_entities=[]forentitiesinentity_list:e=[]# If there are not entities in the textifnotentities.get('denotations'):parsed_entities.append({'text':entities['text'],'text_sha256': hashlib.sha256(entities['text'].encode('utf-8')).hexdigest()})continueforentityinentities['denotations']:other_ids=[idforidinentity['id']ifnotid.startswith("BERN")]entity_type=entity['obj']entity_name=entities['text'][entity['span']['begin']:entity['span']['end']]try:entity_id=[idforidinentity['id']ifid.startswith("BERN")][0]exceptIndexError:entity_id=entity_namee.append({'entity_id': entity_id,'other_ids': other_ids,'entity_type': entity_type,'entity': entity_name})parsed_entities.append({'entities':e,'text':entities['text'],'text_sha256': hashlib.sha256(entities['text'].encode('utf-8')).hexdigest()})
I’ve inspected the results of the named entity linking, and as expected, it is not perfect. For example, it does not identify stem cells as a medical concept. On the other hand, it detected a single entity named “heart, brain, nerves, and kidney”. However, BERN is still the best open-source biomedical model I could find during my investigation.
Construct a knowledge graph
Before looking at relation extraction techniques, we will construct a biomedical knowledge graph using only entities and examine the possible applications. As mentioned, I have prepared a Google Colab notebook that you can use to follow the code examples in this post. To store our graph, we will be using Neo4j. You don’t have to deal with preparing a local Neo4j environment. Instead, you can use a free Neo4j Sandbox instance.
Start the Blank project in the sandbox and copy the connection details to the Colab notebook.
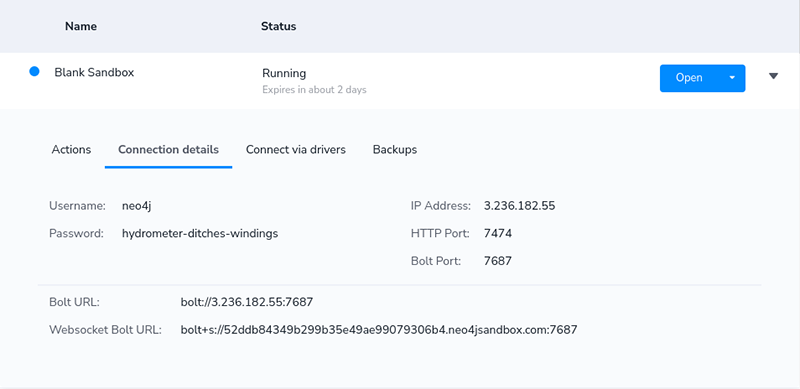
Now you can go ahead and prepare the Neo4j connection in the notebook.
fromneo4jimportGraphDatabaseimportpandas as pdhost='bolt://3.236.182.55:7687'user='neo4j'password='hydrometer-ditches-windings'driver=GraphDatabase.driver(host,auth=(user, password))defneo4j_query(query, params=None):with driver.session() as session:result=session.run(query, params)returnpd.DataFrame([r.values()forrinresult], columns=result.keys())
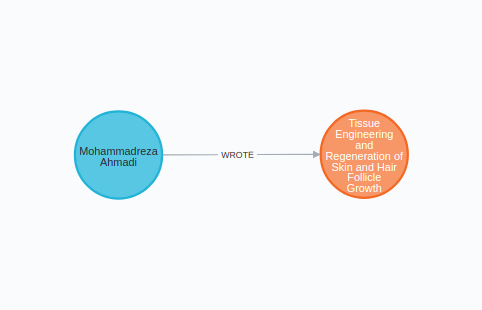
We will start by importing the author and the article into the graph. The article node will contain only the title.
author=article_txt.split("\n")[0]title=" ".join(article_txt.split("\n")[2:4])neo4j_query("""MERGE (a:Author{name:$author})MERGE (b:Article{title:$title})MERGE (a)-[:WROTE]->(b)""", {'title':title,'author':author})
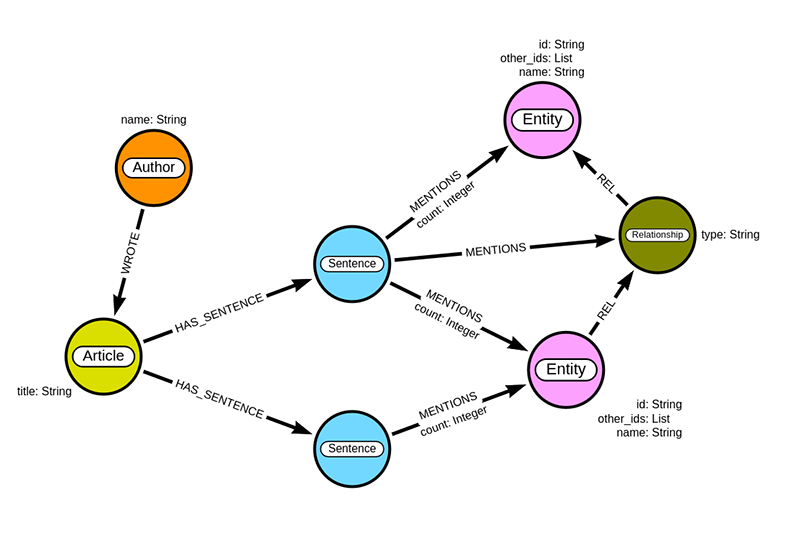
If you open the Neo4j Browser, you should see the following graph.
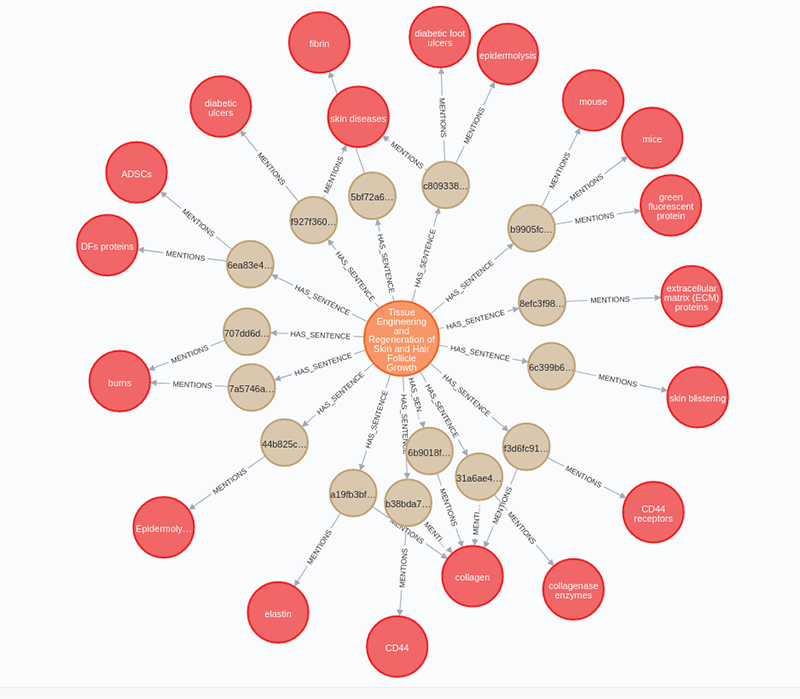
You can import the sentences and mentioned entities by executing the following Cypher query:
neo4j_query("""MATCH (a:Article)UNWIND $data as rowMERGE (s:Sentence{id:row.text_sha256})SET s.text = row.textMERGE (a)-[:HAS_SENTENCE]->(s)WITH s, row.entities as entitiesUNWIND entities as entityMERGE (e:Entity{id:entity.entity_id})ON CREATE SET e.other_ids = entity.other_ids,e.name = entity.entity,e.type = entity.entity_typeMERGE (s)-[m:MENTIONS]->(e)ON CREATE SET m.count = 1ON MATCH SET m.count = m.count + 1""", {'data': parsed_entities})
You can execute the following Cypher query to inspect the constructed graph:
MATCH p=(a:Article)-[:HAS_SENTENCE]->()-[:MENTIONS]->(e:Entity)
RETURN p LIMIT 25If you have correctly imported the data, you should see a similar visualization.
Knowledge graph applications
Even without the relation extraction flow, there are already a couple of use-cases for our graph.
Search engine
We could use our graph as a search engine. For example, you could use the following Cypher query to find sentences or articles that mention a specific medical entity.
MATCH (e:Entity)<-[:MENTIONS]-(s:Sentence)
WHERE e.name = "autoimmune diseases"
RETURN s.text as resultResults

Co-occurrence analysis
The second option is the co-occurrence analysis. You could define co-occurrence between medical entities if they appear in the same sentence or article. I’ve found an article[2] that uses the medical co-occurrence network to predict new possible connections between medical entities.
Link prediction in a MeSH co-occurrence network: preliminary results – PubMed
You could use the following Cypher query to find entities that often co-occur in the same sentence.
MATCH (e1:Entity)<-[:MENTIONS]-()-[:MENTIONS]->(e2:Entity)
MATCH (e1:Entity)<-[:MENTIONS]-()-[:MENTIONS]->(e2:Entity)
WHERE id(e1) < id(e2)
RETURN e1.name as entity1,
e2.name as entity2,
count(*) as cooccurrence
ORDER BY cooccurrence
DESC LIMIT 3Results
| entity1 | entity2 | cooccurrence |
|---|---|---|
| skin diseases | diabetic ulcers | 2 |
| chronic wounds | diabetic ulcers | 2 |
| skin diseases | chronic wounds | 2 |
Obviously, the results would be better if we analyzed thousands or more articles.
Inspect author expertise
You could also use this graph to find the author’s expertise by examining the medical entities they most frequently write about. With this information, you could also suggest future collaborations.
Execute the following Cypher query to inspect which medical entities our single author mentioned in the research paper.
MATCH (a:Author)-[:WROTE]->()-[:HAS_SENTENCE]->()-[:MENTIONS]->(e:Entity)
RETURN a.name as author,
e.name as entity,
MATCH (a:Author)-[:WROTE]->()-[:HAS_SENTENCE]->()-[:MENTIONS]->(e:Entity)
RETURN a.name as author,
e.name as entity,
count(*) as count
ORDER BY count DESC
LIMIT 5Results
| author | entity | count |
|---|---|---|
| Mohammadreza Ahmadi | collagen | 9 |
| Mohammadreza Ahmadi | burns | 4 |
| Mohammadreza Ahmadi | skin diseases | 4 |
| Mohammadreza Ahmadi | collagenase enzymes | 2 |
| Mohammadreza Ahmadi | Epidermolysis bullosa | 2 |
Relation extraction
Now we will try to extract relations between medical concepts. From my experience, the relation extraction is at least an order of magnitude harder than named entity extraction. If you shouldn’t expect perfect results with named entity linking, then you can definitely expect some mistakes with the relation extraction technique.
I’ve been looking for available biomedical relation extraction models but found nothing that works out of the box or doesn’t require fine-tuning. It seems that the field of relation extraction is at the cutting edge, and hopefully, we’ll see more attention about it in the future. Unfortunately, I’m not an NLP expert, so I avoided fine-tuning my own model. Instead, we will be using the zero-shot relation extractor based on the paper Exploring the zero-shot limit of FewRel[3]. While I wouldn’t recommend to put this model into production, it is good enough for a simple demonstration. The model is available on HuggingFace, so we don’t have to deal with training or setting up the model.
fromtransformersimportAutoTokenizerfromzero_shot_reimportRelTaggerModel, RelationExtractormodel=RelTaggerModel.from_pretrained("fractalego/fewrel-zero-shot")tokenizer=AutoTokenizer.from_pretrained("bert-large-uncased-whole-word-masking-finetuned-squad")relations=['associated','interacts']extractor=RelationExtractor(model, tokenizer, relations)
With the zero-shot relation extractor, you can define which relations you would like to detect. In this example, I’ve used the associated and interacts relationships. I’ve also tried more specific relationship types such as treats, causes, and others, but the results were not great.
With this model, you have to define between which pairs of entities you would like to detect relationships. We will use the results of the named entity linking as an input to the relation extraction process. First, we find all the sentences where two or more entities are mentioned and then run them through the relation extraction model to extract any connections. I’ve also defined a threshold value of 0.85, meaning that if a model predicts a link between entities with a probability lower than 0.85, we’ll ignore the prediction.
importitertools# Candidate sentence where there is more than a single entity presentcandidates=[sforsinparsed_entitiesif(s.get('entities'))and(len(s['entities']) >1)]predicted_rels=[]forcincandidates:combinations=itertools.combinations([{'name':x['entity'],'id':x['entity_id']}forxinc['entities']],2)forcombinationinlist(combinations):try:ranked_rels=extractor.rank(text=c['text'].replace(","," "), head=combination[0]['name'], tail=combination[1]['name'])# Define threshold for the most probable relationifranked_rels[0][1] >0.85:predicted_rels.append({'head': combination[0]['id'],'tail': combination[1]['id'],'type':ranked_rels[0][0],'source': c['text_sha256']})except:pass# Store relations to Neo4jneo4j_query("""UNWIND $data as rowMATCH (source:Entity {id: row.head})MATCH (target:Entity {id: row.tail})MATCH (text:Sentence {id: row.source})MERGE (source)-[:REL]->(r:Relation {type: row.type})-[:REL]->(target)MERGE (text)-[:MENTIONS]->(r)""", {'data': predicted_rels})
We store the relationships as well as the source text used to extract that relationship in the graph.
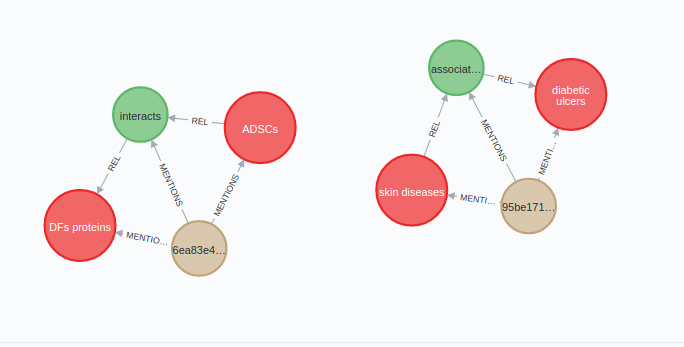
You can examine the extracted relationships between entities and the source text with the following Cypher query:
MATCH (s:Entity)-[:REL]->(r:Relation)-[:REL]->(t:Entity),
(r)<-[:MENTIONS]-(st:Sentence)
RETURN s.name as source_entity,
t.name as target_entity,
r.type as type,
st.text as source_textResults

As mentioned, the NLP model I’ve used to extract relations is not perfect, and since I am not a medical doctor, I don’t know how many connections it missed. However, the ones it detected seem reasonable.
External database enrichment
As I mentioned before, we can still use the external databases like CHEBI or MESH to enrich our graph. For example, our graph contains a medical entity Epidermolysis bullosa and we also know its MeSH id.
You can retrieve the MeSH id of Epidermolysis bullosa with the following query:
MATCH (e:Entity)
WHERE e.name = "Epidermolysis bullosa"
RETURN e.name as entity, e.other_ids as other_idsYou could go ahead and inspect the MeSH to find available information:
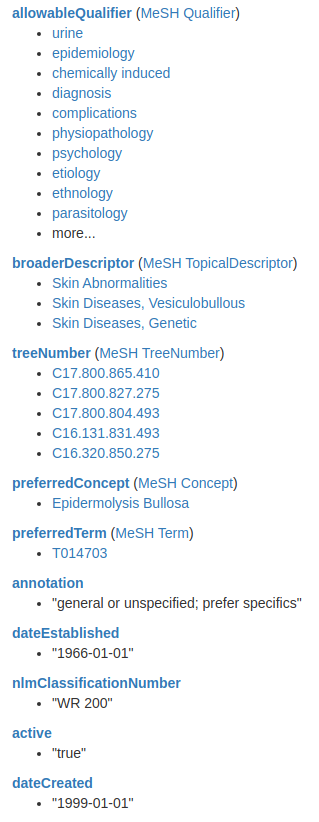
Here is a screenshot of the available information on the MeSH website for Epidermolysis bullosa. As mentioned, I’m not a medical doctor, so I don’t know exactly what would be the best way to model this information in a graph. However, I’ll show you how to retrieve this information in Neo4j using the apoc.load.json procedure to fetch the information from the MeSH REST endpoint. And then, you can ask a domain expert to help you model this information.
The Cypher query to fetch the information from MeSH REST endpoint is:
MATCH (e:Entity)
WHERE e.name = "Epidermolysis bullosa"
WITH e,
[id in e.other_ids WHERE id contains "MESH" | split(id,":")[1]][0] as meshId
CALL apoc.load.json("https://id.nlm.nih.gov/mesh/lookup/details?descriptor=" + meshId) YIELD value
RETURN valueKnowledge graph as machine learning data input
As a final thought, I will quickly walk you through how you could use the biomedical knowledge graph as an input to a machine learning workflow. In recent years, there has been a lot of research and advancement in the node embedding field. Node embedding models translate the network topology into embedding space.
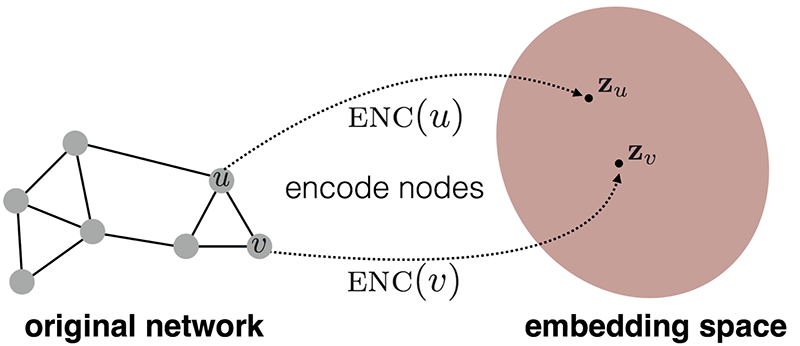
Suppose you constructed a biomedical knowledge graph containing medical entities and concepts, their relations, and enrichment from various medical databases. You could use node embedding techniques to learn the node representations, which are fixed-length vectors, and input them into your machine learning workflow. Various applications are using this approach ranging from drug repurposing to drug side or adverse effect predictions. I’ve found a research paper that uses link prediction for potential treatments of new diseases[4].
Conclusion
The biomedical domain is a prime example where knowledge graphs are applicable. There are many applications ranging from simple search engines to more complicated machine learning workflows. Hopefully, by reading this blog post, you came up with some ideas on how you could use biomedical knowledge graphs to support your applications. You can start a free Neo4j Sandbox and start exploring today.
As always, the code is available on GitHub.
References
[1] D. Kim et al., “A Neural Named Entity Recognition and Multi-Type Normalization Tool for Biomedical Text Mining,” in IEEE Access, vol. 7, pp. 73729–73740, 2019, doi: 10.1109/ACCESS.2019.2920708.
[2] Kastrin A, Rindflesch TC, Hristovski D. Link prediction in a MeSH co-occurrence network: preliminary results. Stud Health Technol Inform. 2014;205:579–83. PMID: 25160252.
[3] Cetoli, A. (2020). Exploring the zero-shot limit of FewRel. In Proceedings of the 28th International Conference on Computational Linguistics (pp. 1447–1451). International Committee on Computational Linguistics.
[4] Zhang, R., Hristovski, D., Schutte, D., Kastrin, A., Fiszman, M., & Kilicoglu, H. (2021). Drug repurposing for COVID-19 via knowledge graph completion. Journal of Biomedical Informatics, 115, 103696.
This article was originally published on Towards Data Science and re-published to TOPBOTS with permission from the author.
Enjoy this article? Sign up for more AI updates.
We’ll let you know when we release more technical education.

Thanks for this extensive tutorial. I am running the colab file shared by you, I am getting the following error while parsing the file “PDFPageCountError: Unable to get page count.” Could you please help on this. Thanks!